Strengthening Pandemic Preparedness
Lessons from SARS-CoV-2 serosurveillance strategies
In May 2023, more than three years after its original declaration, the World Health Organization (WHO) finally ended COVID-19’s status as a global health emergency. Though the pandemic lingers in our collective memory, it must also serve as a crucial reminder for the need to learn from global responses to bolster future pandemic preparedness. Identifying strategies to strengthen the response chain – from pathogen identification to mass vaccination and serosurveillance – is imperative, if we are to prevent future outbreaks from escalating into devastating pandemics.
Although infectious disease outbreaks are inevitable, real-time discovery, monitoring, and control of such events is paramount for guiding public health decision-making. Particularly, serosurveillance during COVID-19 proved to be a valuable tool for identifying emerging variants, monitoring virus transmission rates, and providing a comprehensive understanding of population immunity.
Keeping tabs
Widespread global circulation of SARS-CoV-2 gave rise to new variants that posed a significant threat to public health systems – possessing mutations that enhanced transmissibility and virulence while potentially compromising the efficacy of vaccines, diagnostic tests, and therapeutic measures. As healthcare providers and policymakers navigated these challenges, monitoring the immune status of the population and closely tracking the evolution of variants became essential. Omicron, for example, was found to be capable of evading immunity from previous infection and vaccination, highlighting the importance of routine variant monitoring (1).
Evaluating the presence of neutralizing antibodies and longevity of antibody response helped quantify the efficacy of SARS-CoV-2 vaccines against various variants. Typically, neutralizing antibodies prevent viral entry into human cells by blocking the binding of the SARS-CoV-2 S1 receptor-binding domain (RBD) to the angiotensin-converting enzyme 2 (ACE2) receptor. However, this is not the case for all antibodies, especially those that bind to variants such as Omicron, which have multiple mutations in the RBD subunit – enabling partial escape from antibody neutralization (2,3).
Antibody neutralization assays allow researchers to monitor the efficacy of COVID-19 vaccines and compare it with the naturally occurring antibody response to emerging variants. By keeping up with emerging variants and evaluating the immune response, policymakers and public health officials could make informed decisions and respond effectively to evolving threats.
Evaluating immune response
Given that about 80 percent of SARS-CoV-2 infections result in asymptomatic or mild cases, transmission by asymptomatic and presymptomatic individuals plays a significant role in its spread. Relying solely on monitoring symptomatic cases overlooks a large portion of the population with either current or previous SARS-CoV-2 infection, as well as variations in vaccine and booster timelines. Employing a range of surveillance tools in seroprevalence studies enabled researchers to understand the spread of COVID-19 within the general population – assessing the antibody response and protective immunity resulting from natural infection or vaccination in both symptomatic and asymptomatic individuals. This approach not only helped manage the COVID-19 pandemic, but will also aid in formulating response strategies for future outbreaks or pandemics.
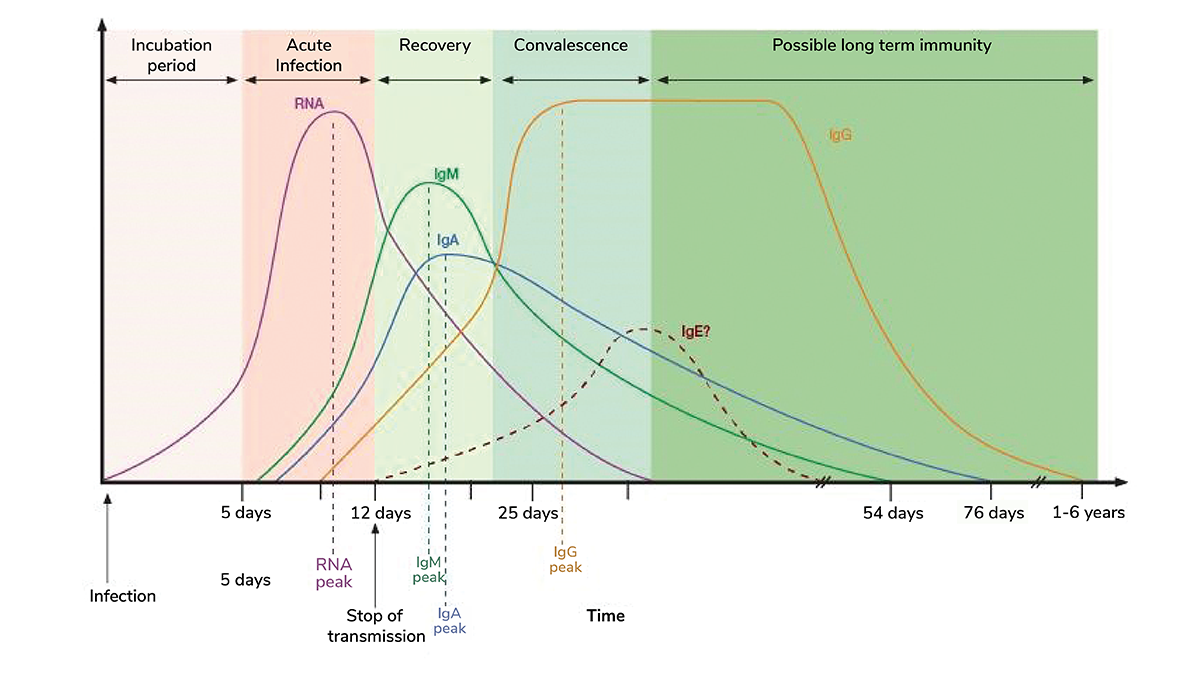
In this case, multiplex serology assays offer a comprehensive evaluation of the isotype-specific antibody response to different SARS-CoV-2 proteins – nucleocapsid, RBD, S1, and S2 subunits – simultaneously (4). IgA, produced shortly after exposure, contributes significantly to virus neutralization. Its absence in the early stages of infection may be associated with severe disease progression, vaccine failure, and viral shedding in immunocompromised patients. IgM levels are typically dominant in the early stages of infection, while IgG levels peak weeks after initial exposure (5). Patients with severe COVID-19 have also shown a delayed IgM and IgG response when compared with patients experiencing mild symptoms (6,7).
Understanding the dynamics of these antibody responses aids in managing symptom progression, improving patient outcomes, monitoring vaccine efficacy, and determining the need for additional boosters.
Future thinking
The toll of COVID-19 underscores the need to strengthen global pandemic preparedness. Real-time monitoring the immune status of the population, employing multiplex assays to assess antibody responses for seroprevalence, and tracking the evolution of variants all provide valuable insights for continuously managing COVID-19 and formulating strategies for future outbreaks (8). Timely intervention is crucial; historical evidence demonstrates the difficulty in eradicating infectious diseases once they go global. By adopting comprehensive serosurveillance platforms and staying vigilant, policymakers and public health officials can respond efficiently to the challenges posed by emerging threats and mitigate their potential impact.
References
N Ferguson et al., “Growth, population distribution and immune escape of Omicron in England,” Imperial College London (2021). Available at: bit.ly/44PZJEO.
C Fenwick et al., “A multiplexed high-throughput neutralization assay reveals a lack of activity against multiple variants after SARS-CoV-2 infection,” medRxiv, [Preprint] (2021).
S Cele et al., “Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization,” Nature, 602, 654 (2022). PMID: 35016196.
C Fenwick et al., “Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies,” J Virol, 95, e01828 (2021). Available at: 33144321.
Y Galipeau et al., “Humoral responses and serological assays in SARS-CoV-2 infections,” Front Immunol, 11, 610688 (2020). PMID: 33391281.
L Shen et al., “Delayed specific IgM antibody responses observed among COVID-19 patients with severe progression,” Emerg Microbes Infect, 9, 1096 (2020). PMID: 32476607.
X Liu et al., “Patterns of IgG and IgM antibody response in COVID-19 patients,” Emerg Microbes Infect, 9, 1269 (2020). PMID: 32515684.
V Margan et al., “Evaluating and Understanding the True Spread of COVID-19 through Seroprevalence,” Bioradiations (2022). Available at: bit.ly/46XA2nt.





